Wahi a nā helu helu o Zhiliwo Big Data Platform, ʻo ka awelika o ka hopena hopena o ka haʻi ʻana i nā pepa Foregene SCI i Pepeluali 2022 he kiʻekiʻe e like me 5.8, a ʻo ka hopena hopena kiʻekiʻe loa he 8.947.
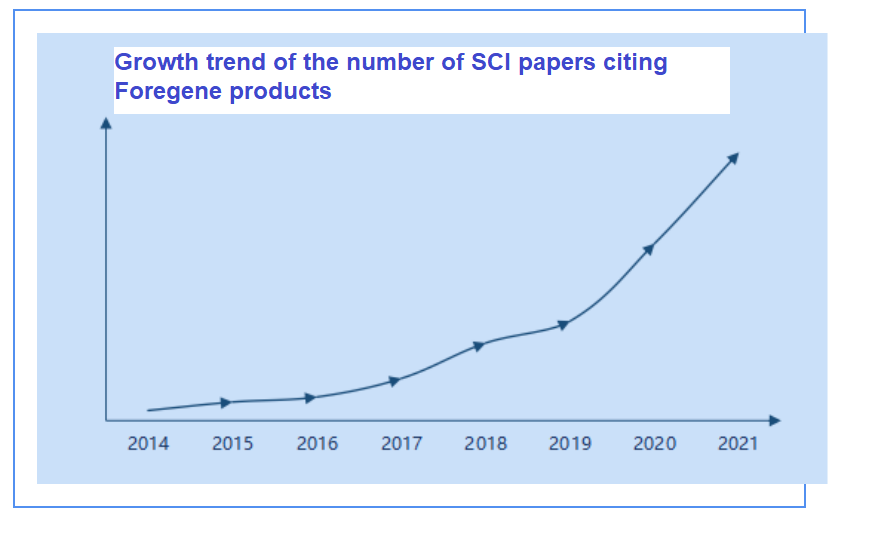
Ma o ka helu helu helu o ka nui o nā pepa SCI o nā huahana Foregene i nā makahiki, ua ʻike ʻia ke ʻano o ka ulu ʻana.
Ua hoʻokumu ʻia ʻo Foregene Co., Ltd. ma 2011. He ʻoihana biotechnology e hoʻohui ana i ka R&D, ka hana, ke kūʻai aku a me ka lawelawe mea kūʻai aku.Mai kona hoʻokumu ʻana, ua kālele ʻo ia i ka hoʻomohala ʻana i nā ʻenehana pili i ka molecular a me ka cell biology pili, nā huahana a me nā lawelawe, a hāʻawi i ka hoʻomaʻemaʻe nucleic acid, ʻike nucleic acid a me nā hopena ʻē aʻe i ka hapa nui o nā mea hoʻohana.ʻO kāna laina huahana noiʻi ʻepekema e pili ana i ka moʻo hoʻomaʻemaʻe RNA, moʻo hoʻomaʻemaʻe DNA, moʻo RT/RT-PCR, moʻo PCR pololei, PCR Mix moʻo, moʻo RT-qPCR pololei a me "Sui" pūʻulu o nā mea molekala pili.Ua hoʻohana nui ʻia nā huahana i ka noiʻi kumu ma nā ʻano ʻepekema ola, lāʻau lapaʻau, mahiʻai, lāʻau lapaʻau, a me ke kaiapuni.Aia nā mea kūʻai ma nā kulanui, nā keʻena noiʻi, nā halemai a me nā ʻāpana ʻē aʻe.
Ua koho ʻo ZhiLiaowo i nā palapala kiʻekiʻe 6 o Foregene i Pepeluali no ka ʻike ʻana o nā mea noiʻi, i mea e ʻike hohonu ai a kamaʻāina i nā huahana Foregene.

Ke poʻo o ka ʻatikala: Hoʻoponopono ka ʻoʻoleʻa substrate i ka hoʻokaʻawale ʻana o nā pūnaewele pluripotent i hoʻokomo ʻia i loko o nā cell endothelial valve puʻuwai.
DOI: 10.1016/j.actbio.2022.02.032
Ka manawa hoʻokuʻu: 2022-2-27
Foregene
Nā Huahana Haʻi ʻia ʻo Literature

Poʻomanaʻo ʻatikala: Ka ʻike ʻana i nā inoa inoa ʻo Mitochondrial Proteome e pili ana me ka Pathogenesis of Pulmonary Arterial Hypertension in the Rat Model.
DOI: 10.1155/2022/8401924
Ka manawa hoʻokuʻu: 2022-2-21
Mea Hoʻohui: Ke Keʻena ʻOihana Lapaʻau, Halemai Hoʻohui Mua o Xinjiang Medical University.
Abstract: ʻO ka Pulmonary arterial hypertension (PAH) kahi maʻi koʻikoʻi a holomua e pili ana i ka puʻuwai a me nā māmā a he pilikia olakino honua e pili ana i nā poʻe a me ke kaiāulu.Ma kēia haʻawina, ua hana maikaʻi ʻia kahi ʻano ʻiole o PAH i hoʻokomo ʻia e ka monocrotaline (MCT).Ua hoʻoholo ʻia nā protein mitochondrial o kēlā me kēia hui o nā ʻiole me ka hoʻohana ʻana i nā ʻenehana proteomic quantitative label-free.Ma muli o ke kūkulu ʻana a me ka hoʻonui ʻana i ka hana o nā pūnaewele protein-protein interaction (PPI), 19 up-regulated mitochondrial genes a me 123 down-regulated mitochondrial genes i nānā ʻia i nā ʻiole PAH.Eia kekahi, i loko o kahi hoʻokolohua hoʻokolohua cohort kūʻokoʻa, ua hōʻike ʻia nā hopena i hōʻike ʻia ʻo 6 up-regulated mitochondrial genes a me 3 down-regulated mitochondrial genes i hōʻike ʻokoʻa ʻia i loko o ka ʻiʻo māmā o nā ʻiole PAH.Ua wānana ʻia nā genes mitochondrial waena o ka miRNA i ka pae transcriptome me ka hoʻohana ʻana i ka waihona RNAInter.Ua ʻike pū ʻia ka haʻawina ʻo bortezomib a me carfilzomib i mau lāʻau lapaʻau no ka mālama ʻana i ka PAH.ʻO ka mea hope loa, hāʻawi kēia haʻawina i ka mālamalama hou i nā biomarkers koʻikoʻi a me nā hoʻolālā therapeutic no nā PAH.
Foregene
Nā Huahana Haʻi ʻia ʻo Literature
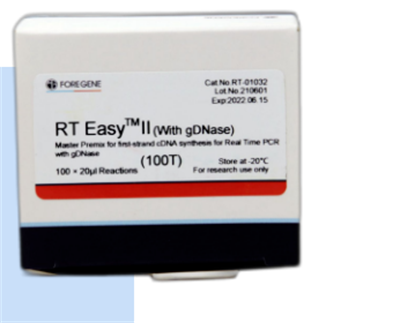
RT Easy II (Master Premix for First Strand cDNA Synthesis No qPCR)
Helu helu: RT-01021
Hōʻike: 50 × 10 μL rxns

Poʻomanaʻo ʻatikala: Hāʻawi ka nalo PTEN i ka naʻau i nā rapalogs i loko o ka maʻi maʻi maʻi maʻi maʻi
DOI: 10.1038/s41401-022-00862-1
Ka manawa hoʻokuʻu: 2022-2-14
Mea Hoʻohui: Ke Keʻena ʻOihana Lapaʻau, Halemai Nanfang, Ke Kulanui Lapaʻau Hema, Guangzhou, Kina
Abstract: ʻO Rapalog kahi mea pale mTORC1 allosteric a he lāʻau lapaʻau i ʻae ʻia no ka maʻi maʻi maʻi maʻi maʻi maʻi maʻemaʻe maʻemaʻe (ccRCC).Hōʻike nā haʻawina e hoʻohana ana i nā laina kelepona ccRCC i loaʻa i nā mea maʻi e hōʻike ana nā cell deficient PTEN i ka hypersensitivity i nā rapalogs.I loko o ka PTEN-deficient ccRCC cell lines, Rapalogs inhibited cell proliferation by inducting G0/G1 hopu me ka hoʻoulu ʻole ʻana i ka apoptosis.Ke hoʻohana nei i nā laina cell isogenic CRISPR/Cas9, ua hōʻoia ka haʻawina i ka hui ma waena o ka holoi ʻana o PTEN a me Rapallo hypersensitivity.I ka hoʻokaʻawale ʻana, ʻaʻole i pili ka holoi ʻana o VHL a i ʻole chromatin-modifying genes (PBRM1, SETD2, BAP1, a i ʻole KDM5C) i nā pane kelepona i nā rapalogs.Ua hōʻike ʻia ka ʻōlelo Ectopic o kahi mutant mTOR i hoʻāla ʻia (C1483F) e kūʻē i ka ulu ʻana o ka ulu ʻana o ka cell PTEN, aʻo ka hoʻokomo ʻana i kahi mutant mTOR (A2034V) i ka lāʻau lapaʻau i ʻae ʻia nā cell ccRCC hemahema PTEN e pakele i nā hopena ulu-suppressive o nā rapalogs.ʻOi aku ka maʻalahi o ka PTEN-deficient ccRCC cell i nā hopena inhibitory o temsirolimus i ka neʻe ʻana o nā cell a me ka ulu ʻana o ka tumora i ka zebrafish a me nā ʻiole xenograft.Ua hōʻike ʻia nā haʻawina e pili ana ka holoi ʻana i ka PTEN me ka rapallo sensitivity a hiki ke hoʻohana ʻia ma ke ʻano he māka no ke koho ʻana i ka lāʻau rapallo i nā maʻi ccRCC.
Foregene
Nā Huahana Haʻi ʻia ʻo Literature

Ke poʻomanaʻo o ka ʻatikala: ʻO ka hana ikaika o ka microbiota o ka wai ʻōpala organik kiʻekiʻe me ka hū paʻakai a me kāna noi.
DOI: 10.1016/j.jece.2022.107377
Ka manawa hoʻokuʻu: 2022-2-12
Mea kākau: School of Life Science, Jiangxi Normal University
Abstract: ʻO ka wai ʻōpala kūlohelohe kiʻekiʻe he wai ʻōpala ʻoihana me nā mea paʻakikī, biodegradability maikaʻi ʻole a me ka mālama biochemical paʻakikī.I mea e hōʻike ai i ka hana intrinsic o ke kaiāulu microbial i ka mālama ʻana i ka wai ʻōpala paʻakai kiʻekiʻe e ka hū paʻakai, ua hana ʻia nā haʻawina ʻike o ke kaiāulu microbial a me nā mea kūlohelohe.Ua hoʻomaikaʻi maikaʻi ʻia ka nui o ka hoʻoneʻe ʻana o ke koi oxygen kemika, NH3–H a me ka nui o ka nitrogen i ka WWTP pilot-scale.Ua hōʻike nā hualoaʻa kiʻekiʻe-throughput sequencing i ka biofortification i hoʻopilikia nui i ka ʻōnaehana kaiāulu microbial.Ma ka pae phylum, ʻoi aku ka nui o nā pūʻulu Proteobacteria a me Ascomycota, ʻoiai ma ka pae genus ka pūʻulu Paracoccus a me Meyeroi.He kuleana koʻikoʻi ʻo Meyerella i ka mālama ʻana i ka wai ʻōpala organik paʻakai kiʻekiʻe, ʻo ia paha ke kumu nui o ka maikaʻi o ka lawe ʻana i nā mea haumia.Eia kekahi, ʻo ka pH, ka oxygen i hoʻoheheʻe ʻia, a me ka leo influent nā kumu nui e pili ana i ka holomua o nā kaiāulu microbial i ka wā o ka bioaugmentation, akā naʻe, ʻaʻole i hoʻopilikia ʻia ka nui o Meyerrotheca e nā ʻano pollutant influent.Ua hōʻike ʻia kēia haʻawina systematically i nā loli ikaika a me nā wānana hana o nā microorganisms i ka wā bioaugmentation, a me ka pilina ma waena o nā kaiāulu microbial a me nā mea pili kaiapuni, e waiho ana i kahi kumu theoretical paʻa no ka hoʻomaʻamaʻa ʻana i ka wai hoʻoneʻe kiʻekiʻe a me ka paʻakai kiʻekiʻe.
Foregene
Nā Huahana Haʻi ʻia ʻo Literature

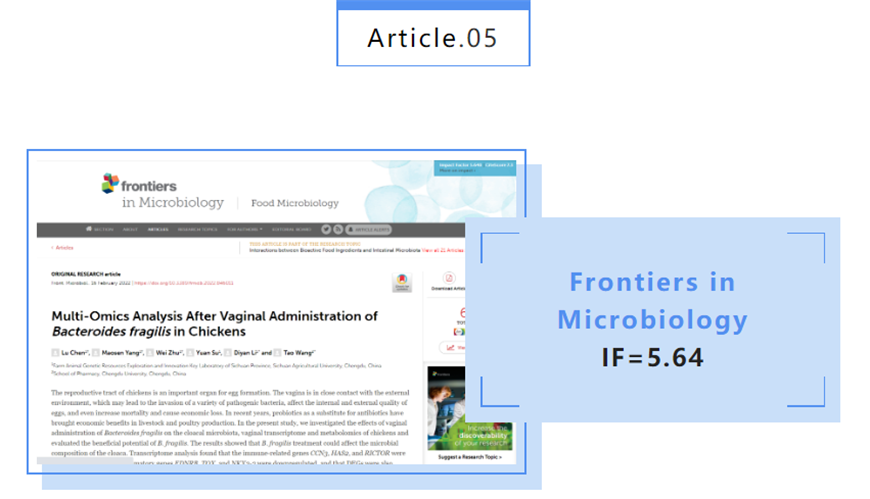
Poʻomanaʻo ʻatikala: ʻIkepili Nui-Omics Ma hope o ka hoʻoponopono ʻana i ka ʻōpū o Bacteroides fragilis i loko o nā moa.
DOI: 10.3389/fmicb.2022.846011
Ka manawa hoʻokuʻu: 2022-2-16
Ka Hoʻohui Mea kākau: Sichuan Key Laboratory of Agricultural and Animal Genetic Resources Development and Innovation
Abstract: ʻO ka ʻaoʻao hānau o nā moa he mea koʻikoʻi no ka hana ʻana i nā hua manu.ʻO ka pilina pili ma waena o ka ʻōpū a me ka ʻāina o waho ke alakaʻi i ka hoʻouka ʻana o nā bacteria pathogenic like ʻole, e hoʻopilikia ana i ka maikaʻi o loko a me waho o nā hua, a me ka hoʻonui ʻana i ka nui o ka make a me ka hoʻokele waiwai.I nā makahiki i hala iho nei, ua lawe mai nā probiotics i nā pōmaikaʻi waiwai i nā holoholona a me ka hana moa ma ke ʻano he ʻokoʻa i nā lāʻau antibiotic.Ua noiʻi ʻia nā hopena o ka hoʻohana ʻana o B. fragilis i ka moa cloacal microbiota, vaginal transcriptome a me ka metabolome e loiloi i ka pono o ka B. fragilis.Ua hōʻike ʻia nā hopena i pili ka mālama ʻana iā B. fragilis i ke ʻano microbial o ka cloaca.Ua ʻike ʻia ka loiloi Transcriptome ua hoʻoponopono ʻia nā genes pili i ka immune CCN3, HAS2, a me RICTOR, a ua hoʻoponopono ʻia nā genes inflammatory EDNRB, TOX, a me NKX2-3, a ua hoʻonui pū ʻia nā DEG i ka hoʻoponopono ʻana o ka mumū, cellular metabolism, a me nā ala pane synaptic.Eia kekahi, pili nui nā metabolites like ʻole me ka biosynthesis hormone steroid, unsaturated fatty acid biosynthesis, a me ka arachidonic acid metabolism, a ua ʻike ʻia nā haʻawina i nā hui ma waena o nā metabolites a me nā genes.I ka hopena, hāʻawi kēia haʻawina i kumu kumu no ka hoʻohana ʻana i ka B. fragilis ma ke ʻano he probiotic hiki ke hana i nā holoholona a me ka hana moa.
Foregene
Nā Huahana Haʻi ʻia ʻo Literature
RT Easy II (Master Premix no ka strand cDNA synthesis mua no qPCR)
Helu helu: RT-01031
Hōʻike: 25 × 20 μL rxns

Ke poʻo o ka ʻatikala: Ke ʻano genetic o Echinococcus granulosus sensu lato ma Kina: Epidemiological study and systematic review
DOI: 10.1111/tbed.14469
Ka manawa hoʻokuʻu: 2022-2-9
Ka Hoʻohui Mea kākau: Sichuan Key Laboratory of Agricultural and Animal Genetic Resources Development and Innovation
Abstract: ʻO Cystic echinococcosis (CE) kahi maʻi zoonotic tropical zoonotic i hoʻokumu ʻia e Echinococcus granulosus a noho mau i ka pilikia olakino ākea honua.Ua ʻike ʻia nā genotypes ʻike ʻia o Echinococcus granulosus ʻē aʻe ma mua o G4 ma Kina, ʻoi aku ka nui o ka Qinghai-Tibet Plateau, a ʻokoʻa ko lākou ʻano genetic ma kēia māhele o ka honua.Wahi a kahi loiloi systematic, ʻo ka genetic composition o E. granulosa ma koʻu ʻāina penei: E. granulosa (G1, G3), 98.3%;Echinococcus intermedius (G5), 0.1%;Echinococcus intermedius (G6, G7), 1.4%;a me Canada Echinococcus (G8, G10), 0.2%.ʻO ke kikoʻī, ʻo ka nui o ka maʻi maʻi G1 he 97.7% a ua ʻike ʻia e kahi ākea ākea ākea a me ka māhele ʻāina.Ua hōʻike ʻia nā hopena epidemiological i kahi haku mele E. coli paʻa a maikaʻi loa i loko o nā hipa a me ka yak mai ʻekolu mau panalāʻau kiʻekiʻe CE (Xinjiang, Sichuan, a me Qinghai).ʻO nā hipa (287/406, 70.7%) i ʻoi aku ka nui o nā cysts momona ma mua o ka yak (28/184, 15.2%).I nā makahiki he 29 i hala iho nei, aia he 51 mau haplotypes Echinococcus granulosus cox1 ma Kina.ʻO ka haplotype o nā kūpuna (Hap_2) i noho mau i ka haplotype maʻamau, 12 mau haplotypes maʻamau i hoʻopau ʻia, a ua ʻike ʻia he 9 mau haplotypes hou i hōʻike ʻia i ka wā o ka loiloi.Hōʻike nā mea i ʻike ʻia ʻo ka hoʻomaʻamaʻa pono ʻana o nā hipa a me ka huakaʻi hoʻokele hoʻokele EG95 o nā yaks e pili pono i ke kūlana genotype o kēia manawa.
Foregene
Nā Huahana Haʻi ʻia ʻo Literature
Hoʻokaʻaʻike:
Maggie |Hoʻomohala ʻoihana
Foregene Co., Ltd
M: +86-15281067355
HKH: www.foreivd.com
Ka manawa kau: Apr-08-2022








